Overview
This tutorial will demonstrate how to adjust for exposure misclassification. First, generate a dataset of 100,000 rows with the following binary variables:
- X = Exposure (1 = exposed, 0 = not exposed)
- Y = Outcome (1 = outcome, 0 = no outcome)
- C = Known Confounder (1 = present, 0 = absent)
- Xstar = Misclassified exposure (1 = exposed, 0 = not exposed)
set.seed(1234)
n <- 100000
C <- rbinom(n, 1, .5)
X <- rbinom(n, 1, plogis(-.5 + .5 * C))
Y <- rbinom(n, 1, plogis(-.5 + log(2) * X + .5 * C))
Xstar <- ifelse(X == 1 & Y == 1, rbinom(n, 1, .75),
(ifelse(X == 1 & Y == 0, rbinom(n, 1, .65),
(ifelse(X == 0 & Y == 1, rbinom(n, 1, .25), rbinom(n, 1, .35))))))
df <- data.frame(X, Xstar, Y, C)
rm(C, X, Y, Xstar)
This data reflects the following causal relationships:
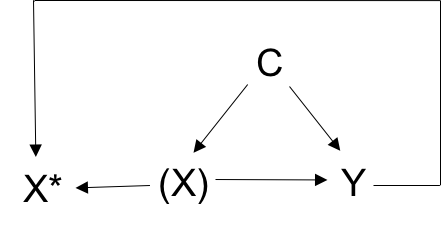
From this dataset note that P(Y=1|X=1, C=c, U=u) / P(Y=1|X=0, C=c, U=u) should equal expit(log(2)). Therefore, odds(Y=1|X=1, C=c, U=u) / odds(Y=1|X=0, C=c, U=u) = ORYX = exp(log(2)) = 2.
Compare the biased (exposure misclassified) model to the bias-free model.
nobias_model <- glm(Y ~ X + C, family = binomial(link = "logit"), data = df)
exp(coef(nobias_model)[2])
c(exp(coef(nobias_model)[2] + summary(nobias_model)$coef[2, 2] * qnorm(.025)),
exp(coef(nobias_model)[2] + summary(nobias_model)$coef[2, 2] * qnorm(.975)))
ORYX = 2.04 (1.99, 2.10)
This estimate corresponds to the odds ratio we would expect based off of the derivation of Y.
biased_model <- glm(Y ~ Xstar + C, family = binomial(link = "logit"), data = df)
exp(coef(biased_model)[2])
c(exp(coef(biased_model)[2] + summary(biased_model)$coef[2, 2] * qnorm(.025)),
exp(coef(biased_model)[2] + summary(biased_model)$coef[2, 2] * qnorm(.975)))
ORYX = 1.27 (1.24, 1.31)
The odds ratio of the effect of X on Y decreases from ~2 to ~1.3 when the exposure X is misclassified in the model. Misclassification is sometimes inevitable due to inaccurate measurement tools or human error. This bias analysis will allow for correct inference when Xstar is used instead of X. We will rely on assumptions of how the misclassified exposure, outcome, and known confounder(s) affect the true exposure (each assumption corresponding to a bias parameter). Using these assumptions, we will create a model to predict the probability of X, which will then be incorporated as a weight in the outcome regression. This regression weight provides the bias adjustment.
Normally, the values of the bias parameters are obtained externally from the literature or from an internal validation sub-study. However, for this proof of concept, we can obtain the exact values of the parameters. We will perform the final regression of Y on Xstar instead of X. Since we know the values of X, we can model how X is affected by Xstar, C, and Y to obtain accurate bias parameters.
x_model <- glm(X ~ Xstar + Y + C, family = binomial(link = "logit"), data = df)
summary(x_model)
These parameters can be interpreted as follows:
- Intercept = log[odds(X=1|Xstar=0, C=0, Y=0)]
- Xstar coefficient = log[odds(X=1|Xstar=1, C=0, Y=0)] / log[odds(X=1|Xstar=0, C=0, Y=0)] i.e. the log odds ratio denoting the amount by which the log odds of X=1 changes for every 1 unit increase in Xstar among the C=0, Y=0 subgroup.
- Y coefficient = log[odds(X=1|Xstar=0, C=0, Y=1)] / log[odds(X=1|Xstar=0, C=0, Y=0)]
- C coefficient = log[odds(X=1|Xstar=0, C=1, Y=0)] / log[odds(X=1|Xstar=0, C=0, Y=0)]
Now that values for the bias parameters have been obtained, we’ll use these values to perform the bias adjustment with two different approaches. In both cases, we’ll build the analysis within a function for quick reiteration. Bootstrapping will be used in order to obtain a confidence interval for the ORYX estimate.
1. Weighting Approach
The steps for the weighting approach are as follows:
- Sample with replacement from the dataset to get the bootstrap sample.
- Predict the probability of X by combining the bias parameters with the data for Xstar, Y, and C via the inverse logit transformation.
- Duplicate the bootstrap dataset and merge these two copies.
- In the combined data, assign variable Xbar, which equals 1 in the first data copy and equals 0 in the second data copy.
- Create variable x_weight, which equals the probability of X=1 in the first copy and equals 1 minus the probability of X=1 in the second copy.
- With the combined dataset, model the weighted logistic outcome regression [P(Y=1)| Xbar, C]. The weight used in this regression comes from x_weight.
- Save the exponentiated Xbar coefficient, corresponding to the odds ratio effect estimate of X on Y.
- Repeat the above steps with a new bootstrap sample.
- With the resulting vector of odds ratio estimates, obtain the final estimate and confidence interval from the median and 2.5, 97.5 quantiles, respectively.
adjust_emc_wgt_loop <- function(
coef_0, coef_xstar, coef_y, coef_c, nreps, plot = FALSE
) {
est <- vector()
for (i in 1:nreps) {
bdf <- df[sample(seq_len(n), n, replace = TRUE), ]
x_probability <- plogis(
coef_0 + coef_xstar * bdf$Xstar + coef_y * bdf$Y + coef_c * bdf$C
)
combined <- dplyr::bind_rows(bdf, bdf)
combined$Xbar <- rep(c(1, 0), each = n)
combined$x_weight <- c(x_probability, 1 - x_probability)
final_model <- glm(Y ~ Xbar + C, family = binomial(link = "logit"),
data = combined, weights = combined$x_weight)
est[i] <- exp(coef(final_model)[2])
}
out <- list(
estimate = round(median(est), 2),
ci = round(quantile(est, c(.025, .975)), 2)
)
if (plot) {
out$hist <- hist(exp(est))
}
return(out)
}
2. Imputation Approach
The steps for the imputation approach are as follows:
- Sample with replacement from the dataset to get the bootstrap sample.
- Predict the probability of X by combining the bias parameters with the data for Xstar, Y, and C via the inverse logit transformation.
- Impute the value of the exposure, Xpred, across Bernoulli trials where the probability of each trial corresponds to the probability of X determined above.
- With the bootstrap sample, model the logistic outcome regression [P(Y=1)| Xpred, C].
- Save the exponentiated Xpred coefficient, corresponding to the odds ratio effect estimate of X on Y.
- Repeat the above steps with a new bootstrap sample.
- With the resulting vector of odds ratio estimates, obtain the final estimate and confidence interval from the median and 2.5, 97.5 quantiles, respectively.
adjust_emc_imp_loop <- function(
coef_0, coef_xstar, coef_y, coef_c, nreps, plot = FALSE
) {
est <- vector()
for (i in 1:nreps) {
bdf <- df[sample(seq_len(n), n, replace = TRUE), ]
bdf$Xpred <- rbinom(
n, 1, plogis(coef_0 + coef_xstar * bdf$Xstar +
coef_y * bdf$Y + coef_c * bdf$C)
)
final_model <- glm(Y ~ Xpred + C,
family = binomial(link = "logit"),
data = bdf)
est[i] <- exp(coef(final_model)[2])
}
out <- list(
estimate = round(median(est), 2),
ci = round(quantile(est, c(.025, .975)), 2)
)
if (plot) {
out$hist <- hist(exp(est))
}
return(out)
}
Evaluate
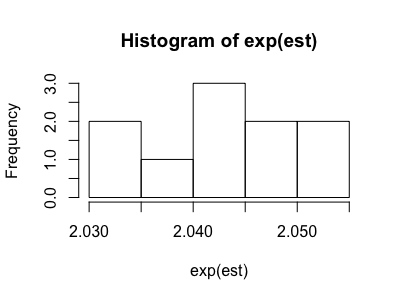
We can run the analysis using different values of the bias parameters. When we use the known, correct values for the bias parameters that we obtained earlier we obtain ORYX = 2.05 (2.04, 2.06), representing the bias-adjusted effect estimate we expect based on the derivation of the data.
set.seed(1234)
correct_results <- adjust_emc_imp_loop(
coef_0 = coef(x_model)[1],
coef_xstar = coef(x_model)[2],
coef_y = coef(x_model)[3],
coef_c = coef(x_model)[4],
nreps = 10
)
correct_results$estimate
correct_results$ci
The output can also include a histogram showing the distribution of the ORYX estimates from each bootstrap sample. We can analyze this plot to see how well the odds ratios converge.
If instead we use bias parameters that are each double the correct value, we obtain ORYX = 2.85 (2.84, 2.88), an incorrect estimate of effect.
set.seed(1234)
incorrect_results <- adjust_emc_imp_loop(
coef_0 = coef(x_model)[1] * 2,
coef_xstar = coef(x_model)[2] * 2,
coef_y = coef(x_model)[3] * 2,
coef_c = coef(x_model)[4] * 2,
nreps = 10
)
incorrect_results$estimate
incorrect_results$ci
Very similar results are obtained from using the imputation approach. But don’t take my word for it, see for yourself! The full code for this analysis is available here.